Note
Go to the end to download the full example code.
1D Biased Double Well¶
Estimation of an overdamped Langevin in presence of biased dynamics.
Euler [ 4.22054548 -12.51555831 13.83274923 -6.30298083 0.48478253
0.52170744 0.49355682 0.5007274 ]
Elerian [ 4.22054795 -12.51555549 13.83275452 -6.30296867 0.48487525
0.52178501 0.49358168 0.50077429]
Kessler [ 4.22055274 -12.51553497 13.83277905 -6.30292702 0.48667153
0.52063655 0.4921984 0.50298026]
Drozdov [ 4.22057785 -12.5155274 13.83278778 -6.3029196 0.48686415
0.52065394 0.49209304 0.50305591]
import numpy as np
import matplotlib.pyplot as plt
import folie as fl
from copy import deepcopy
coeff = 0.2 * np.array([0, 0, -4.5, 0, 0.1])
free_energy = np.polynomial.Polynomial(coeff)
D = np.array([0.5])
drift_coeff = D * np.array([-coeff[1], -2 * coeff[2], -3 * coeff[3], -4 * coeff[4]])
drift_function = fl.functions.Polynomial(deg=3, coefficients=drift_coeff)
diff_function = fl.functions.Polynomial(deg=0, coefficients=D)
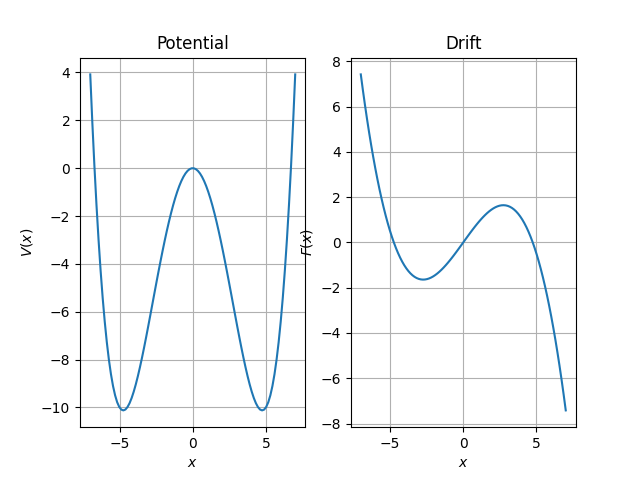
# Plot of Free Energy and Force
x_values = np.linspace(-7, 7, 100)
fig, axs = plt.subplots(1, 2)
axs[0].plot(x_values, free_energy(x_values))
axs[1].plot(x_values, drift_function(x_values.reshape(len(x_values), 1)))
axs[0].set_title("Potential")
axs[0].set_xlabel("$x$")
axs[0].set_ylabel("$V(x)$")
axs[0].grid()
axs[1].set_title("Drift")
axs[1].set_xlabel("$x$")
axs[1].set_ylabel("$F(x)$")
axs[1].grid()
# Define model to simulate and type of simulator to use
model_simu = fl.models.overdamped.Overdamped(drift_function, diffusion=diff_function)
simulator = fl.simulations.ABMD_Simulator(fl.simulations.EulerStepper(model_simu), 1e-3, k=10.0, xstop=6.0)
# initialize positions
ntraj = 30
q0 = np.empty(ntraj)
for i in range(len(q0)):
q0[i] = -6
# Calculate Trajectory
time_steps = 25000
data = simulator.run(time_steps, q0, save_every=1)
xmax = np.concatenate(simulator.xmax_hist, axis=1).T
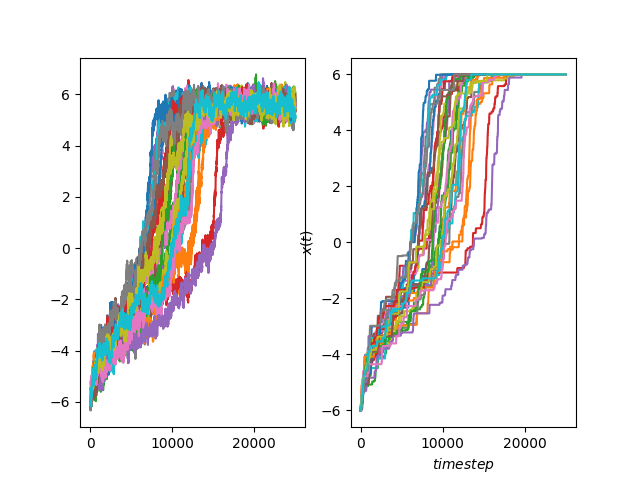
# Plot the resulting trajectories
fig, axs = plt.subplots(1, 2)
for n, trj in enumerate(data):
axs[0].plot(trj["x"])
axs[1].plot(xmax[:, n])
axs[1].set_xlabel("$timestep$")
axs[1].set_ylabel("$x(t)$")
axs[1].grid()
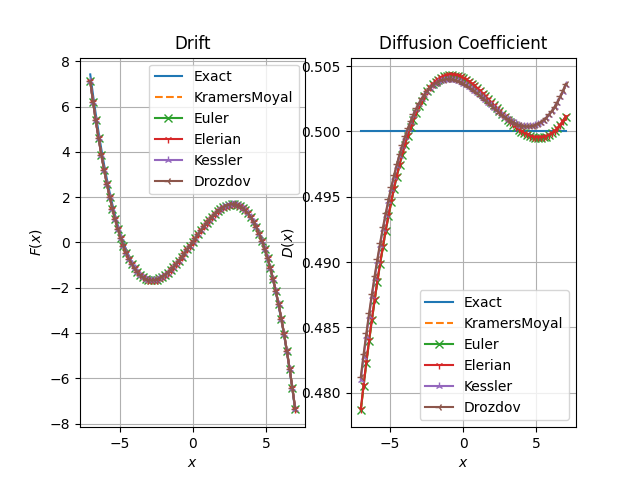
fig, axs = plt.subplots(1, 2)
axs[0].set_title("Drift")
axs[0].set_xlabel("$x$")
axs[0].set_ylabel("$F(x)$")
axs[0].grid()
axs[1].set_title("Diffusion Coefficient") # i think should be diffusion coefficient
axs[1].set_xlabel("$x$")
axs[1].set_ylabel("$D(x)$")
axs[1].grid()
xfa = np.linspace(-7.0, 7.0, 75)
model_simu.remove_bias()
axs[0].plot(xfa, model_simu.drift(xfa.reshape(-1, 1)), label="Exact")
axs[1].plot(xfa, model_simu.diffusion(xfa.reshape(-1, 1)), label="Exact")
domain = fl.MeshedDomain.create_from_range(np.linspace(data.stats.min, data.stats.max, 4).ravel())
trainmodel = fl.models.Overdamped(fl.functions.BSplinesFunction(domain), has_bias=True)
name = "KramersMoyal"
estimator = fl.KramersMoyalEstimator(trainmodel)
res = estimator.fit_fetch(data)
res.remove_bias()
axs[0].plot(xfa, res.drift(xfa.reshape(-1, 1)), "--", label=name)
axs[1].plot(xfa, res.diffusion(xfa.reshape(-1, 1)), "--", label=name)
for name, marker, transitioncls in zip(
["Euler", "Elerian", "Kessler", "Drozdov"],
["x", "1", "2", "3"],
[
fl.EulerDensity,
fl.ElerianDensity,
fl.KesslerDensity,
fl.DrozdovDensity,
],
):
trainmodel = fl.models.Overdamped(fl.functions.BSplinesFunction(domain), has_bias=True)
estimator = fl.LikelihoodEstimator(transitioncls(trainmodel), n_jobs=4)
res = estimator.fit_fetch(deepcopy(data))
print(name, res.coefficients)
res.remove_bias()
axs[0].plot(xfa, res.drift(xfa.reshape(-1, 1)), marker=marker, label=name)
axs[1].plot(xfa, res.diffusion(xfa.reshape(-1, 1)), marker=marker, label=name)
axs[0].legend()
axs[1].legend()
plt.show()
Total running time of the script: (0 minutes 32.082 seconds)