Note
Go to the end to download the full example code.
2D Double Well¶
Estimation of an overdamped Langevin.
import numpy as np
import matplotlib.pyplot as plt
import folie as fl
from mpl_toolkits.mplot3d import Axes3D
from copy import deepcopy
""" Script for simulation of 2D double well and projection along user provided direction, No fitting is carried out """
x = np.linspace(-1.8, 1.8, 36)
y = np.linspace(-1.8, 1.8, 36)
input = np.transpose(np.array([x, y]))
D = 0.5
diff_function = fl.functions.Polynomial(deg=0, coefficients=D * np.eye(2, 2))
a, b = 5, 10
drift_quartic2d = fl.functions.Quartic2D(a=D * a, b=D * b) # simple way to multiply D*Potential here force is the SDE force (meandispl) ## use this when you need the drift ###
quartic2d = fl.functions.Quartic2D(a=a, b=b) # Real potential , here force is just -grad pot ## use this when you need the potential energy ###
X, Y = np.meshgrid(x, y)
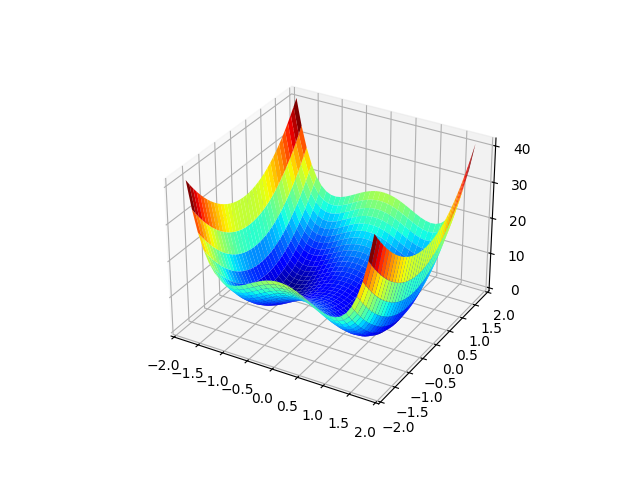
# Plot potential surface
pot = quartic2d.potential_plot(X, Y)
fig = plt.figure()
ax = plt.axes(projection="3d")
ax.plot_surface(X, Y, pot, rstride=1, cstride=1, cmap="jet", edgecolor="none")
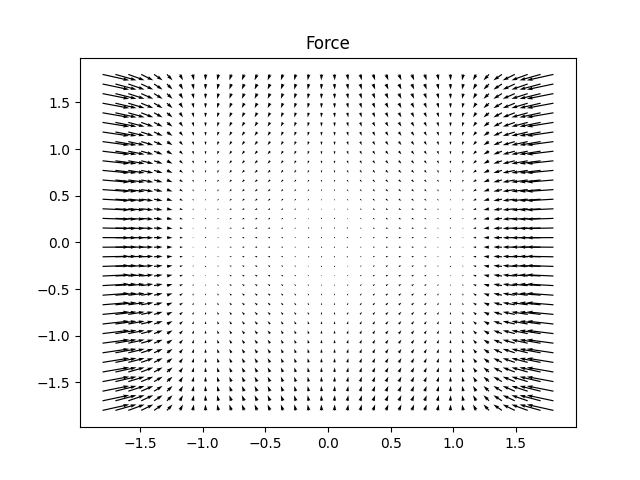
# Plot Force function
ff = quartic2d.force(input) # returns x and y components of the force : x_comp =ff[:,0] , y_comp =ff[:,1]
U, V = np.meshgrid(ff[:, 0], ff[:, 1])
fig, ax = plt.subplots()
ax.quiver(x, y, U, V)
ax.set_title("Force")
dt = 5e-4
model_simu = fl.models.overdamped.Overdamped(drift_quartic2d, diffusion=diff_function)
simulator = fl.simulations.Simulator(fl.simulations.EulerStepper(model_simu), dt)
# initialize positions
ntraj = 30
q0 = np.empty(shape=[ntraj, 2])
for i in range(ntraj):
for j in range(2):
q0[i][j] = 0.000
# Calculate Trajectory
time_steps = 3000
data = simulator.run(time_steps, q0, save_every=1)
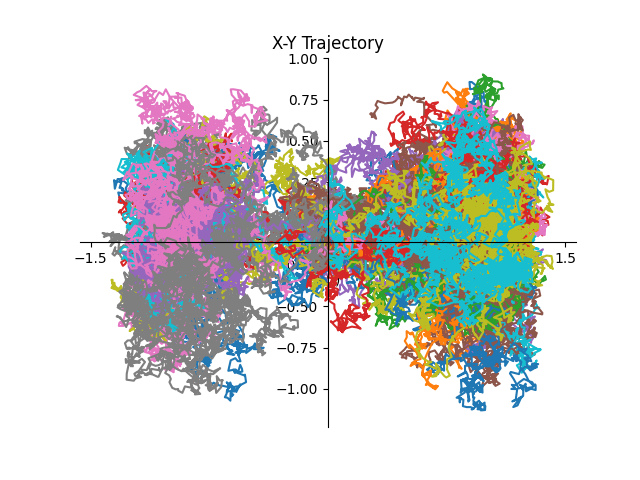
# Plot the resulting trajectories
fig, axs = plt.subplots()
for n, trj in enumerate(data):
axs.plot(trj["x"][:, 0], trj["x"][:, 1])
axs.spines["left"].set_position("center")
axs.spines["right"].set_color("none")
axs.spines["bottom"].set_position("center")
axs.spines["top"].set_color("none")
axs.xaxis.set_ticks_position("bottom")
axs.yaxis.set_ticks_position("left")
axs.set_xlabel("$X(t)$")
axs.set_ylabel("$Y(t)$")
axs.set_title("X-Y Trajectory")
axs.grid()
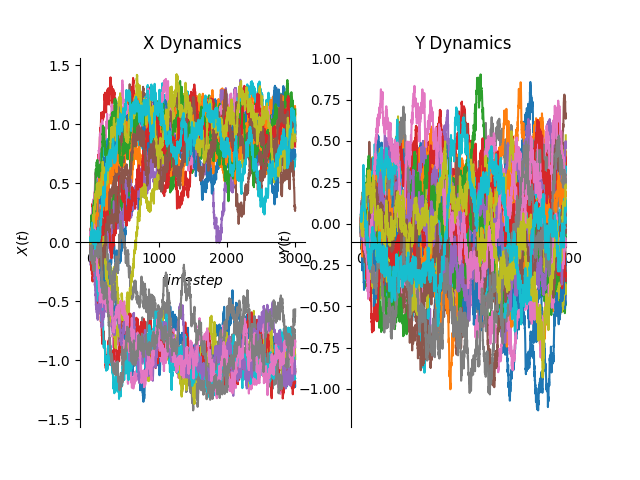
# plot Trajectories
fig, bb = plt.subplots(1, 2)
for n, trj in enumerate(data):
bb[0].plot(trj["x"][:, 0])
bb[1].plot(trj["x"][:, 1])
# Set visible axis
bb[0].spines["right"].set_color("none")
bb[0].spines["bottom"].set_position("center")
bb[0].spines["top"].set_color("none")
bb[0].xaxis.set_ticks_position("bottom")
bb[0].yaxis.set_ticks_position("left")
bb[0].set_xlabel("$timestep$")
bb[0].set_ylabel("$X(t)$")
# Set visible axis
bb[1].spines["right"].set_color("none")
bb[1].spines["bottom"].set_position("center")
bb[1].spines["top"].set_color("none")
bb[1].xaxis.set_ticks_position("bottom")
bb[1].yaxis.set_ticks_position("left")
bb[1].set_xlabel("$timestep$")
bb[1].set_ylabel("$Y(t)$")
bb[0].set_title("X Dynamics")
bb[1].set_title("Y Dynamics")
PROJECTION ALONG CHOSEN COORDINATE #¶
# Choose unit versor of direction
u = np.array([1, 1])
u_norm = (1 / np.linalg.norm(u, 2)) * u
w = np.empty_like(trj["x"][:, 0])
proj_data = fl.Trajectories(dt=1e-3)
fig, axs = plt.subplots()
for n, trj in enumerate(data):
for i in range(len(trj["x"])):
w[i] = np.dot(trj["x"][i], u_norm)
proj_data.append(fl.Trajectory(1e-3, deepcopy(w.reshape(len(trj["x"][:, 0]), 1))))
axs.plot(proj_data[n]["x"])
axs.set_xlabel("$timesteps$")
axs.set_ylabel("$w(t)$")
axs.set_title("trajectory projected along $u =$" + str(u) + " direction")
axs.grid()
plt.show()
![trajectory projected along $u =$[1 1] direction](../../_images/sphx_glr_plot_2D_Double_Well_005.png)
Total running time of the script: (0 minutes 1.403 seconds)